GWAS prephasing and imputation
Posted on February 20, 2017
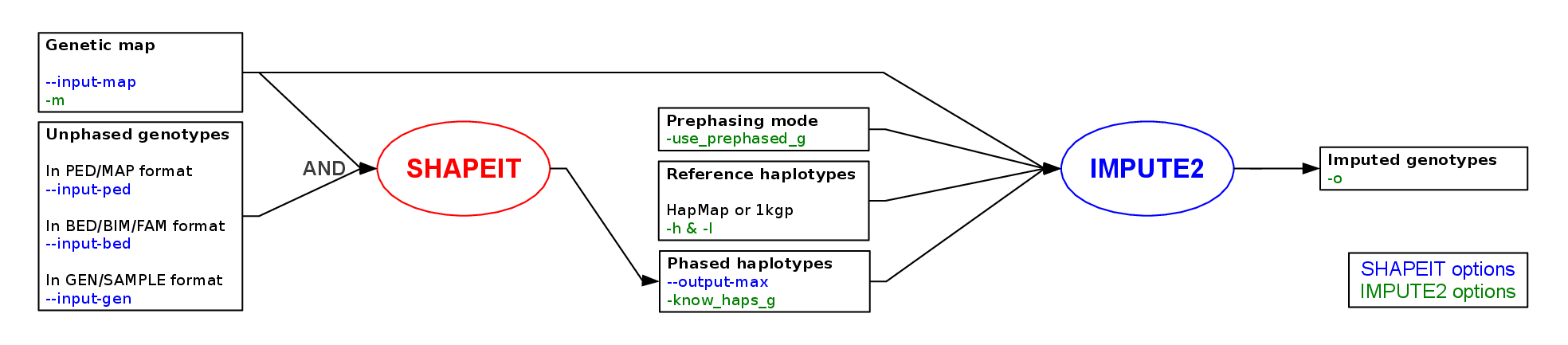
Below shows a general workflow for carrying out a GWAS prephasing and imputation using 1000GP phase3. In this guide, I will focus on the processing of GWAS imputation in a detailed manner.
Background
A major use of phasing is haplotype estimation of GWAS samples in order to speed up imputation from large reference panel of haplotypes such as 1000 Genomes. The current recommendation is that GWAS samples are first ‘pre-phased’ using the most accurate method available. The subsequent imputation step (which involves imputing alleles from one set of haplotypes into another set) is fast. As new haplotype reference sets become available imputation can be re-run much more efficiently. The approach we recommend is:
Step1: Alignment of the SNPs
SNP positions in build 37
The most recent 1,000 genomes haplotypes are defined at SNPs that use build37 coordinates. You have thus to make sure that your GWAS SNPs use also the same version. If it is not the case, you can use the UCSC liftOver tool to perform the conversion to build37 coordinates
Strand alignment
This is a crucial step of prephasing/imputation to make sure that the GWAS dataset is well aligned with the reference panel of haplotypes. Correcting strand A/T and G/C SNPs is a big concern. Genotype Harmonizer is an easy to use tool helping you accomplish this job.
Assume we have study gwas file and 1000 genome refernce file in PLINK format.
wget http://www.molgenis.org/downloads/GenotypeHarmonizer/GenotypeHarmonizer-1.4.20-dist.tar.gz # download Genotype Harmonizer
mkdir alignment
java -Xmx40g -jar /user/path/to/GenotypeHarmonizer.jar \
--inputType PLINK_BED \
--input path_to_study_gwas \ # PLINK file prefix only
--update-id \
--outputType PLINK_BED \
--output alignment/all_chrs \
--refType PLINK_BED \
--ref path_to_reference # PLINK file prefix onlyIt will generate harmonized all_chrs.bed all_chrs.bim and all_chrs.fam in the alignment folder. Genotype Harmonizer uses Linkage disequilibrium (LD) patterns to determine the correct strand G/C and A/T SNPs.
Step2: Phasing the GWAS samples
Once you GWAS dataset correctly aligned to the reference panel, we strongly recommend to phase each chromosome in a single run instead of making chunks. It makes the procedure much easier and increase downstream imputation quality.
wget http://mathgen.stats.ox.ac.uk/genetics_software/shapeit/old_versions/shapeit.v2.r790.Ubuntu_12.04.4.static.tar.gz # download shapeit2 executable file
wget https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.tgz # download reference haplotypes, genetic maps files are also included
wget https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3_chrX.tgz # download reference haplotypesYou can write a small BASH script to run the phasing.
!/usr/bin/env bash
PLINK="/path/to/plink2"
SHAPEIT="path/to/shapeit2"
for i in {1..22}
do
mkdir chr$i
$PLINK --bfile path/to/all_chrs --chr $i --make-bed --out ./chr$i/unphased_chr$i
$SHAPEIT -B ./chr${i}/unphased_chr${i} -M ./references/genetic_map_chr${i}_combined_b37.txt -O ./chr${i}/phased_chr${i} -T 10
echo "phased_chr$i generated!"
doneNotes on clusters
Suppose that you want to prephase your GWAS on a cluster where each node has X CPU cores. In this case, the approach we recommend is:
- To reserve a complete cluster node for each SHAPEIT job
- To run each SHAPEIT job with X threads to fully load the CPU-cores of a node
Step3: Imputation of the GWAS samples
Once SHAPEIT has produced haplotype estimates, you can use IMPUTE2 to impute untyped genotypes using the latest release of the 1000 Genomes haplotypes.
download impute2
wget https://mathgen.stats.ox.ac.uk/impute/impute_v2.3.2_x86_64_static.tgz # download impute2 executable fileExample
impute2 -use_prephased_g -Ne 20000 -iter 30 -align_by_maf_g -os 0 1 2 3 -seed 1000000 -o_gz -int 1 5000001 -h 1000GP_Phase3_chr22.hap.gz -l 1000GP_Phase3_chr22.legend.gz -m genetic_map_chr22_combined_b37.txt -known_haps_g phased_chr22.haps -o chr22.chunk1 # chr22.chunk1.gz generated
impute2 -use_prephased_g -Ne 20000 -iter 30 -align_by_maf_g -os 0 1 2 3 -seed 1000000 -o_gz -int 5000001 10000001 -h 1000GP_Phase3_chr22.hap.gz -l 1000GP_Phase3_chr22.legend.gz -m genetic_map_chr22_combined_b37.txt -known_haps_g phased_chr22.haps -o chr22.chunk2 # chr22.chunk2.gz generatedSeveral comments on the previous command line:
- Prephased GWAS haplotypes are specified using -known_haps_g
- The flag -use_prephased_g is used to set IMPUTE2 in the prephasing mode
- The option -int 5000001 10000001 is used to specify the region to be imputed. Combine all the chunks
cat chr22.chunk1.gz chr22.chunk2.gz chr22.chunk3.gz > chr22_chunkAll.gen.gzX chromosome prephasing and imputation
Step 1: Prephasing using SHAPEIT
shapeit -B chrX.unphased -M chrX.gmap.gz -O chrX.phased --chrXStep 2: Imputation using IMPUTE2
impute2 -chrX -use_prephased_g -known_haps_g chrX.phased.haps -sample_known_haps_g chrX.phased.sample -h chrX.reference.hap.gz -l chrX.reference.legend.gz -m chrX.gmap.gz -o chrX.imputed -int 10e6 11e6Two comments:
- You must use the -chrX flag for IMPUTE2 to proceed with X chromosome imputation
- You must give the SAMPLE file generated by SHAPEIT to IMPUTE2. This SAMPLE has a sex column that gives the gender of the GWAS individuals.